diff --git a/README.md b/README.md
index 9f123ebe..d781b999 100644
--- a/README.md
+++ b/README.md
@@ -9,13 +9,13 @@
diff --git a/docs/en/datasets/explorer/api.md b/docs/en/datasets/explorer/api.md
index 37f5262d..33ba47e6 100644
--- a/docs/en/datasets/explorer/api.md
+++ b/docs/en/datasets/explorer/api.md
@@ -300,8 +300,6 @@ You can also visualize the embedding space using the plotting tool of your choic
```python
import matplotlib.pyplot as plt
-import numpy as np
-from mpl_toolkits.mplot3d import Axes3D
from sklearn.decomposition import PCA
# Reduce dimensions using PCA to 3 components for visualization in 3D
diff --git a/docs/en/guides/isolating-segmentation-objects.md b/docs/en/guides/isolating-segmentation-objects.md
index 3885e624..e8c227b2 100644
--- a/docs/en/guides/isolating-segmentation-objects.md
+++ b/docs/en/guides/isolating-segmentation-objects.md
@@ -14,20 +14,7 @@ After performing the [Segment Task](../tasks/segment.md), it's sometimes desirab
## Recipe Walk Through
-1. Begin with the necessary imports
-
- ```python
- from pathlib import Path
-
- import cv2
- import numpy as np
-
- from ultralytics import YOLO
- ```
-
- ???+ tip "Ultralytics Install"
-
- See the Ultralytics [Quickstart](../quickstart.md/#install-ultralytics) Installation section for a quick walkthrough on installing the required libraries.
+1. See the [Ultralytics Quickstart Installation section](../quickstart.md/#install-ultralytics) for a quick walkthrough on installing the required libraries.
***
@@ -61,6 +48,10 @@ After performing the [Segment Task](../tasks/segment.md), it's sometimes desirab
3. Now iterate over the results and the contours. For workflows that want to save an image to file, the source image `base-name` and the detection `class-label` are retrieved for later use (optional).
```{ .py .annotate }
+ from pathlib import Path
+
+ import numpy as np
+
# (2) Iterate detection results (helpful for multiple images)
for r in res:
img = np.copy(r.orig_img)
@@ -86,6 +77,8 @@ After performing the [Segment Task](../tasks/segment.md), it's sometimes desirab
{ width="240", align="right" }
```{ .py .annotate }
+ import cv2
+
# Create binary mask
b_mask = np.zeros(img.shape[:2], np.uint8)
@@ -178,12 +171,11 @@ After performing the [Segment Task](../tasks/segment.md), it's sometimes desirab
Additional steps required to crop image to only include object region.
{ align="right" }
- ``` { .py .annotate }
+ ```{ .py .annotate }
# (1) Bounding box coordinates
x1, y1, x2, y2 = c.boxes.xyxy.cpu().numpy().squeeze().astype(np.int32)
# Crop image to object region
iso_crop = isolated[y1:y2, x1:x2]
-
```
1. For more information on bounding box results, see [Boxes Section from Predict Mode](../modes/predict.md/#boxes)
@@ -225,12 +217,11 @@ After performing the [Segment Task](../tasks/segment.md), it's sometimes desirab
Additional steps required to crop image to only include object region.
{ align="right" }
- ``` { .py .annotate }
+ ```{ .py .annotate }
# (1) Bounding box coordinates
x1, y1, x2, y2 = c.boxes.xyxy.cpu().numpy().squeeze().astype(np.int32)
# Crop image to object region
iso_crop = isolated[y1:y2, x1:x2]
-
```
1. For more information on bounding box results, see [Boxes Section from Predict Mode](../modes/predict.md/#boxes)
diff --git a/docs/en/guides/kfold-cross-validation.md b/docs/en/guides/kfold-cross-validation.md
index b18ab3fd..2c75da2e 100644
--- a/docs/en/guides/kfold-cross-validation.md
+++ b/docs/en/guides/kfold-cross-validation.md
@@ -57,25 +57,13 @@ Without further ado, let's dive in!
## Generating Feature Vectors for Object Detection Dataset
-1. Start by creating a new Python file and import the required libraries.
-
- ```python
- import datetime
- import shutil
- from collections import Counter
- from pathlib import Path
-
- import numpy as np
- import pandas as pd
- import yaml
- from sklearn.model_selection import KFold
-
- from ultralytics import YOLO
- ```
+1. Start by creating a new `example.py` Python file for the steps below.
2. Proceed to retrieve all label files for your dataset.
```python
+ from pathlib import Path
+
dataset_path = Path("./Fruit-detection") # replace with 'path/to/dataset' for your custom data
labels = sorted(dataset_path.rglob("*labels/*.txt")) # all data in 'labels'
```
@@ -92,6 +80,8 @@ Without further ado, let's dive in!
4. Initialize an empty `pandas` DataFrame.
```python
+ import pandas as pd
+
indx = [l.stem for l in labels] # uses base filename as ID (no extension)
labels_df = pd.DataFrame([], columns=cls_idx, index=indx)
```
@@ -99,6 +89,8 @@ Without further ado, let's dive in!
5. Count the instances of each class-label present in the annotation files.
```python
+ from collections import Counter
+
for label in labels:
lbl_counter = Counter()
@@ -142,6 +134,8 @@ The rows index the label files, each corresponding to an image in your dataset,
- By setting `random_state=M` where `M` is a chosen integer, you can obtain repeatable results.
```python
+ from sklearn.model_selection import KFold
+
ksplit = 5
kf = KFold(n_splits=ksplit, shuffle=True, random_state=20) # setting random_state for repeatable results
@@ -178,6 +172,8 @@ The rows index the label files, each corresponding to an image in your dataset,
4. Next, we create the directories and dataset YAML files for each split.
```python
+ import datetime
+
supported_extensions = [".jpg", ".jpeg", ".png"]
# Initialize an empty list to store image file paths
@@ -222,6 +218,8 @@ The rows index the label files, each corresponding to an image in your dataset,
- **NOTE:** The time required for this portion of the code will vary based on the size of your dataset and your system hardware.
```python
+ import shutil
+
for image, label in zip(images, labels):
for split, k_split in folds_df.loc[image.stem].items():
# Destination directory
@@ -247,6 +245,8 @@ fold_lbl_distrb.to_csv(save_path / "kfold_label_distribution.csv")
1. First, load the YOLO model.
```python
+ from ultralytics import YOLO
+
weights_path = "path/to/weights.pt"
model = YOLO(weights_path, task="detect")
```
diff --git a/docs/en/guides/sahi-tiled-inference.md b/docs/en/guides/sahi-tiled-inference.md
index 4305f66c..e9287b3c 100644
--- a/docs/en/guides/sahi-tiled-inference.md
+++ b/docs/en/guides/sahi-tiled-inference.md
@@ -60,12 +60,6 @@ pip install -U ultralytics sahi
Here's how to import the necessary modules and download a YOLOv8 model and some test images:
```python
-from pathlib import Path
-
-from IPython.display import Image
-from sahi import AutoDetectionModel
-from sahi.predict import get_prediction, get_sliced_prediction, predict
-from sahi.utils.cv import read_image
from sahi.utils.file import download_from_url
from sahi.utils.yolov8 import download_yolov8s_model
@@ -91,6 +85,8 @@ download_from_url(
You can instantiate a YOLOv8 model for object detection like this:
```python
+from sahi import AutoDetectionModel
+
detection_model = AutoDetectionModel.from_pretrained(
model_type="yolov8",
model_path=yolov8_model_path,
@@ -104,6 +100,8 @@ detection_model = AutoDetectionModel.from_pretrained(
Perform standard inference using an image path or a numpy image.
```python
+from sahi.predict import get_prediction
+
# With an image path
result = get_prediction("demo_data/small-vehicles1.jpeg", detection_model)
@@ -125,6 +123,8 @@ Image("demo_data/prediction_visual.png")
Perform sliced inference by specifying the slice dimensions and overlap ratios:
```python
+from sahi.predict import get_sliced_prediction
+
result = get_sliced_prediction(
"demo_data/small-vehicles1.jpeg",
detection_model,
@@ -155,6 +155,8 @@ result.to_fiftyone_detections()[:3]
For batch prediction on a directory of images:
```python
+from sahi.predict import predict
+
predict(
model_type="yolov8",
model_path="path/to/yolov8n.pt",
diff --git a/docs/en/guides/security-alarm-system.md b/docs/en/guides/security-alarm-system.md
index 5ed15e1c..a8c83567 100644
--- a/docs/en/guides/security-alarm-system.md
+++ b/docs/en/guides/security-alarm-system.md
@@ -27,22 +27,6 @@ The Security Alarm System Project utilizing Ultralytics YOLOv8 integrates advanc
### Code
-#### Import Libraries
-
-```python
-import smtplib
-from email.mime.multipart import MIMEMultipart
-from email.mime.text import MIMEText
-from time import time
-
-import cv2
-import numpy as np
-import torch
-
-from ultralytics import YOLO
-from ultralytics.utils.plotting import Annotator, colors
-```
-
#### Set up the parameters of the message
???+ tip "Note"
@@ -60,6 +44,8 @@ to_email = "" # receiver email
#### Server creation and authentication
```python
+import smtplib
+
server = smtplib.SMTP("smtp.gmail.com: 587")
server.starttls()
server.login(from_email, password)
@@ -68,6 +54,10 @@ server.login(from_email, password)
#### Email Send Function
```python
+from email.mime.multipart import MIMEMultipart
+from email.mime.text import MIMEText
+
+
def send_email(to_email, from_email, object_detected=1):
"""Sends an email notification indicating the number of objects detected; defaults to 1 object."""
message = MIMEMultipart()
@@ -84,6 +74,15 @@ def send_email(to_email, from_email, object_detected=1):
#### Object Detection and Alert Sender
```python
+from time import time
+
+import cv2
+import torch
+
+from ultralytics import YOLO
+from ultralytics.utils.plotting import Annotator, colors
+
+
class ObjectDetection:
def __init__(self, capture_index):
"""Initializes an ObjectDetection instance with a given camera index."""
@@ -109,7 +108,7 @@ class ObjectDetection:
def display_fps(self, im0):
"""Displays the FPS on an image `im0` by calculating and overlaying as white text on a black rectangle."""
self.end_time = time()
- fps = 1 / np.round(self.end_time - self.start_time, 2)
+ fps = 1 / round(self.end_time - self.start_time, 2)
text = f"FPS: {int(fps)}"
text_size = cv2.getTextSize(text, cv2.FONT_HERSHEY_SIMPLEX, 1.0, 2)[0]
gap = 10
diff --git a/docs/en/guides/triton-inference-server.md b/docs/en/guides/triton-inference-server.md
index cc1c5b33..6b9b496b 100644
--- a/docs/en/guides/triton-inference-server.md
+++ b/docs/en/guides/triton-inference-server.md
@@ -126,7 +126,7 @@ Then run inference using the Triton Server model:
from ultralytics import YOLO
# Load the Triton Server model
-model = YOLO(f"http://localhost:8000/yolo", task="detect")
+model = YOLO("http://localhost:8000/yolo", task="detect")
# Run inference on the server
results = model("path/to/image.jpg")
diff --git a/docs/en/guides/view-results-in-terminal.md b/docs/en/guides/view-results-in-terminal.md
index ba99e3a2..2a2fedc4 100644
--- a/docs/en/guides/view-results-in-terminal.md
+++ b/docs/en/guides/view-results-in-terminal.md
@@ -18,7 +18,7 @@ When connecting to a remote machine, normally visualizing image results is not p
!!! warning
- Only compatible with Linux and MacOS. Check the VSCode [repository](https://github.com/microsoft/vscode), check [Issue status](https://github.com/microsoft/vscode/issues/198622), or [documentation](https://code.visualstudio.com/docs) for updates about Windows support to view images in terminal with `sixel`.
+ Only compatible with Linux and MacOS. Check the [VSCode repository](https://github.com/microsoft/vscode), check [Issue status](https://github.com/microsoft/vscode/issues/198622), or [documentation](https://code.visualstudio.com/docs) for updates about Windows support to view images in terminal with `sixel`.
The VSCode compatible protocols for viewing images using the integrated terminal are [`sixel`](https://en.wikipedia.org/wiki/Sixel) and [`iTerm`](https://iterm2.com/documentation-images.html). This guide will demonstrate use of the `sixel` protocol.
@@ -31,28 +31,17 @@ The VSCode compatible protocols for viewing images using the integrated terminal
"terminal.integrated.enableImages": false
```
-
- 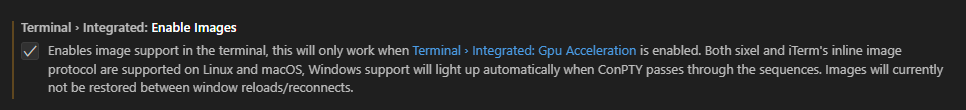 -
-
+
+  +
+
-1. Install the `python-sixel` library in your virtual environment. This is a [fork](https://github.com/lubosz/python-sixel?tab=readme-ov-file) of the `PySixel` library, which is no longer maintained.
+2. Install the `python-sixel` library in your virtual environment. This is a [fork](https://github.com/lubosz/python-sixel?tab=readme-ov-file) of the `PySixel` library, which is no longer maintained.
```bash
pip install sixel
```
-1. Import the relevant libraries
-
- ```py
- import io
-
- import cv2 as cv
- from sixel import SixelWriter
-
- from ultralytics import YOLO
- ```
-
-1. Load a model and execute inference, then plot the results and store in a variable. See more about inference arguments and working with results on the [predict mode](../modes/predict.md) page.
+3. Load a model and execute inference, then plot the results and store in a variable. See more about inference arguments and working with results on the [predict mode](../modes/predict.md) page.
```{ .py .annotate }
from ultralytics import YOLO
@@ -69,11 +58,15 @@ The VSCode compatible protocols for viewing images using the integrated terminal
1. See [plot method parameters](../modes/predict.md#plot-method-parameters) to see possible arguments to use.
-1. Now, use OpenCV to convert the `numpy.ndarray` to `bytes` data. Then use `io.BytesIO` to make a "file-like" object.
+4. Now, use OpenCV to convert the `numpy.ndarray` to `bytes` data. Then use `io.BytesIO` to make a "file-like" object.
```{ .py .annotate }
+ import io
+
+ import cv2
+
# Results image as bytes
- im_bytes = cv.imencode(
+ im_bytes = cv2.imencode(
".png", # (1)!
plot,
)[1].tobytes() # (2)!
@@ -85,9 +78,11 @@ The VSCode compatible protocols for viewing images using the integrated terminal
1. It's possible to use other image extensions as well.
2. Only the object at index `1` that is returned is needed.
-1. Create a `SixelWriter` instance, and then use the `.draw()` method to draw the image in the terminal.
+5. Create a `SixelWriter` instance, and then use the `.draw()` method to draw the image in the terminal.
+
+ ```python
+ from sixel import SixelWriter
- ```py
# Create sixel writer object
w = SixelWriter()
@@ -110,7 +105,7 @@ The VSCode compatible protocols for viewing images using the integrated terminal
```{ .py .annotate }
import io
-import cv2 as cv
+import cv2
from sixel import SixelWriter
from ultralytics import YOLO
@@ -125,7 +120,7 @@ results = model.predict(source="ultralytics/assets/bus.jpg")
plot = results[0].plot() # (3)!
# Results image as bytes
-im_bytes = cv.imencode(
+im_bytes = cv2.imencode(
".png", # (1)!
plot,
)[1].tobytes() # (2)!
diff --git a/docs/en/guides/vision-eye.md b/docs/en/guides/vision-eye.md
index e60fc31e..c5ade6fd 100644
--- a/docs/en/guides/vision-eye.md
+++ b/docs/en/guides/vision-eye.md
@@ -116,7 +116,7 @@ keywords: VisionEye, YOLOv8, Ultralytics, object mapping, object tracking, dista
import cv2
from ultralytics import YOLO
- from ultralytics.utils.plotting import Annotator, colors
+ from ultralytics.utils.plotting import Annotator
model = YOLO("yolov8s.pt")
cap = cv2.VideoCapture("Path/to/video/file.mp4")
diff --git a/docs/en/hub/inference-api.md b/docs/en/hub/inference-api.md
index 68f9ef9e..8357e607 100644
--- a/docs/en/hub/inference-api.md
+++ b/docs/en/hub/inference-api.md
@@ -28,7 +28,7 @@ To access the [Ultralytics HUB](https://bit.ly/ultralytics_hub) Inference API us
import requests
# API URL, use actual MODEL_ID
-url = f"https://api.ultralytics.com/v1/predict/MODEL_ID"
+url = "https://api.ultralytics.com/v1/predict/MODEL_ID"
# Headers, use actual API_KEY
headers = {"x-api-key": "API_KEY"}
@@ -117,7 +117,7 @@ The [Ultralytics HUB](https://bit.ly/ultralytics_hub) Inference API returns a JS
import requests
# API URL, use actual MODEL_ID
- url = f"https://api.ultralytics.com/v1/predict/MODEL_ID"
+ url = "https://api.ultralytics.com/v1/predict/MODEL_ID"
# Headers, use actual API_KEY
headers = {"x-api-key": "API_KEY"}
@@ -185,7 +185,7 @@ The [Ultralytics HUB](https://bit.ly/ultralytics_hub) Inference API returns a JS
import requests
# API URL, use actual MODEL_ID
- url = f"https://api.ultralytics.com/v1/predict/MODEL_ID"
+ url = "https://api.ultralytics.com/v1/predict/MODEL_ID"
# Headers, use actual API_KEY
headers = {"x-api-key": "API_KEY"}
@@ -257,7 +257,7 @@ The [Ultralytics HUB](https://bit.ly/ultralytics_hub) Inference API returns a JS
import requests
# API URL, use actual MODEL_ID
- url = f"https://api.ultralytics.com/v1/predict/MODEL_ID"
+ url = "https://api.ultralytics.com/v1/predict/MODEL_ID"
# Headers, use actual API_KEY
headers = {"x-api-key": "API_KEY"}
@@ -331,7 +331,7 @@ The [Ultralytics HUB](https://bit.ly/ultralytics_hub) Inference API returns a JS
import requests
# API URL, use actual MODEL_ID
- url = f"https://api.ultralytics.com/v1/predict/MODEL_ID"
+ url = "https://api.ultralytics.com/v1/predict/MODEL_ID"
# Headers, use actual API_KEY
headers = {"x-api-key": "API_KEY"}
@@ -400,7 +400,7 @@ The [Ultralytics HUB](https://bit.ly/ultralytics_hub) Inference API returns a JS
import requests
# API URL, use actual MODEL_ID
- url = f"https://api.ultralytics.com/v1/predict/MODEL_ID"
+ url = "https://api.ultralytics.com/v1/predict/MODEL_ID"
# Headers, use actual API_KEY
headers = {"x-api-key": "API_KEY"}
diff --git a/docs/en/modes/predict.md b/docs/en/modes/predict.md
index 1fb021dc..0f99456c 100644
--- a/docs/en/modes/predict.md
+++ b/docs/en/modes/predict.md
@@ -249,8 +249,6 @@ Below are code examples for using each source type:
Run inference on a collection of images, URLs, videos and directories listed in a CSV file.
```python
- import torch
-
from ultralytics import YOLO
# Load a pretrained YOLOv8n model
diff --git a/docs/en/usage/callbacks.md b/docs/en/usage/callbacks.md
index e88ffac8..5a03ef56 100644
--- a/docs/en/usage/callbacks.md
+++ b/docs/en/usage/callbacks.md
@@ -41,7 +41,7 @@ def on_predict_batch_end(predictor):
# Create a YOLO model instance
-model = YOLO(f"yolov8n.pt")
+model = YOLO("yolov8n.pt")
# Add the custom callback to the model
model.add_callback("on_predict_batch_end", on_predict_batch_end)
diff --git a/docs/en/usage/simple-utilities.md b/docs/en/usage/simple-utilities.md
index e68508da..3619eef3 100644
--- a/docs/en/usage/simple-utilities.md
+++ b/docs/en/usage/simple-utilities.md
@@ -349,6 +349,16 @@ from ultralytics.utils.ops import (
xyxy2ltwh, # xyxy → top-left corner, w, h
xyxy2xywhn, # pixel → normalized
)
+
+for func in (
+ ltwh2xywh,
+ ltwh2xyxy,
+ xywh2ltwh,
+ xywh2xyxy,
+ xywhn2xyxy,
+ xyxy2ltwh,
+ xyxy2xywhn):
+ print(help(func)) # print function docstrings
```
See docstring for each function or visit the `ultralytics.utils.ops` [reference page](../reference/utils/ops.md) to read more about each function.
@@ -467,7 +477,10 @@ Want or need to use the formats of [images or videos types supported](../modes/p
from ultralytics.data.utils import IMG_FORMATS, VID_FORMATS
print(IMG_FORMATS)
-# >>> ('bmp', 'dng', 'jpeg', 'jpg', 'mpo', 'png', 'tif', 'tiff', 'webp', 'pfm')
+# {'tiff', 'pfm', 'bmp', 'mpo', 'dng', 'jpeg', 'png', 'webp', 'tif', 'jpg'}
+
+print(VID_FORMATS)
+# {'avi', 'mpg', 'wmv', 'mpeg', 'm4v', 'mov', 'mp4', 'asf', 'mkv', 'ts', 'gif', 'webm'}
```
### Make Divisible
 -
-  +
+  +
+ 
 -
-  +
+  +
+ 
-
+